Table Of Content

The above tips are also applicable to a slightly different form of PCR called quantitative reverse transcription PCR (RT-qPCR). RT-qPCR allows you to perform PCR starting from RNA instead of DNA. If your RNA has been stored in TRIzol® or another similar reagent, learn more about Zymo Research’s Direct-zol RNA Purification Kits. These kits are made to efficiently purify total RNA that is suitable for RT-qPCR from TRI Reagent®, eliminating the need for chloroform, phase separation or precipitation steps. This specifies the max amplicon size for a PCR target to be detected by Primer-BLAST. In general, the non-specific targets become less of a concern if their sizes are very large since PCR is much less efficient for larger amplicons.
Using custom-built primers and nanopore sequencing to evaluate CO-utilizer bacterial and archaeal populations linked ... - Nature.com
Using custom-built primers and nanopore sequencing to evaluate CO-utilizer bacterial and archaeal populations linked ....
Posted: Mon, 09 Oct 2023 07:00:00 GMT [source]
IVT Product Purification
The value of this parameter is less than the actual concentration of oligos in the reaction because it is the concentration of annealing oligos, which in turn depends on the amount of template (including PCR product) in a given cycle. This concentration increases a great deal during a PCR; fortunately PCR seems quite robust for a variety of oligo melting temperatures. The minimal number of contiguous nucleotide base matches between the query sequence and the target sequence that is needed for BLAST to detect the targets. Set a lower value if you need to find target sequences with more mismatches to your primers. Oligonucleotide primers are necessary when running a PCR reaction.
Additives That Benefit G-C Rich Templates
Primer design may start in silico, but it is ultimately proven at the bench. Here are the top recommendations for how to design primers for PCR and real time quantitative PCR (qPCR), reverse transcription quantitative PCR (RT-qPCR), bisulfite PCR, and methylation specific PCR (MSP). You can choose to exclude sequences in the selected database from specificity checking if you are not concerned about these. There are a large number of predicted Refseq transcripts in the Refseq mRNA, Refseq RNA and nr database. There are also many uncultured/environmental sample sequencesare in the nr database. This specifies the minimal number of bases that the primer must anneal to the template at 5' side (i.e., toward start of the primer) or 3' side (i.e., toward end of the primer) of the exon-exon junction.
Necessity for the design of a reference sequence
The forward primer creates copies of the 5’-3’ strand whereas the reverse primer makes copies of the complementary (runs 3’-5’) strand. To solve this type of problem in DNA replication or PCR experiment, primers are used, allowing the DNA polymerase to attach even in the middle of the lagging strand. The area replicated between two primers is called an Okazaki fragment. It is important that you do a quick consistency check -- you want to avoid having the added 5´ and 3´ flanking sequences base pair with the region of interest (ROI).
For cases where users submit a pre-existing primer pair to perform specificity checking (the primer-only case), an artificial template sequence is generated for the BLAST search by connecting both primers with a 20 base spacer region of N’s. This ensures that each primer will be treated separately in the BLAST search and thus achieves the equivalent effect of performing a separate BLAST search for each primer. To further minimize the search time, all regions on the template that do not contain candidate primers are masked out to avoid irrelevant BLAST hits. Since all candidate primer locations on the template are established by Primer3 already, amplification targets (amplicons) for all primer pairs can be identified using the single BLAST result above. A primer pair is deemed to be specific only if it has no amplicons on any targets other than the submitted template within the specificity checking threshold specified by the user.
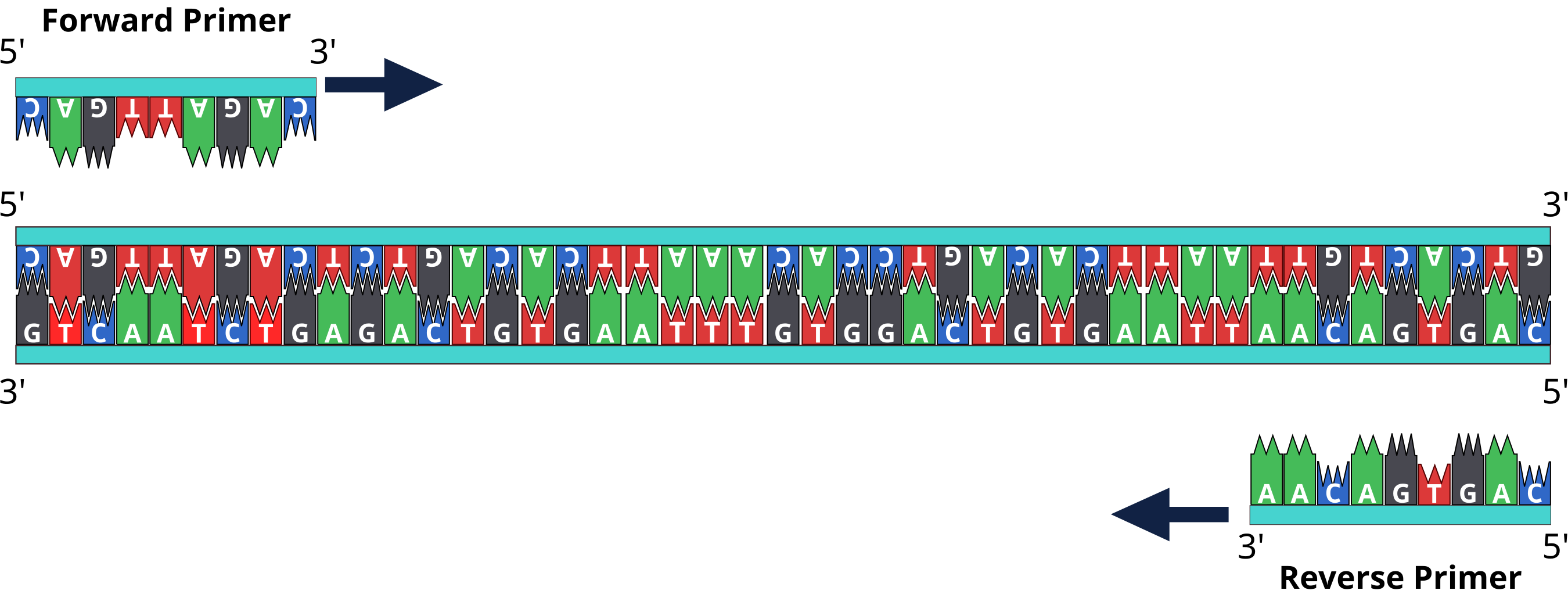
Choosing appropriate primers is probably the single most important factor affecting the polymerase chain reaction (PCR). Specific amplification of the intended target requires that primers do not have matches to other targets in certain orientations and within certain distances that allow undesired amplification. The process of designing specific primers typically involves two stages. First, the primers flanking regions of interest are generated either manually or using software tools; then they are searched against an appropriate nucleotide sequence database using tools such as BLAST to examine the potential targets.
A One-Step PCR-Based Assay to Evaluate the Efficiency and Precision of Genomic DNA-Editing Tools - ScienceDirect.com
A One-Step PCR-Based Assay to Evaluate the Efficiency and Precision of Genomic DNA-Editing Tools.
Posted: Sun, 03 Dec 2017 08:42:38 GMT [source]
Sequence selected for designing forward primer (text highlighted in blue that is pointed by arrowhead). The red box indicates the parameters of the selected sequence like Tm, length of the sequence, and GC%. You can observe several parameters of the selected sequence from the header section of the tools (red box). The parameters are updated in live, so you can select or deselect the nucleotides to match the required criteria. The primer design steps remain the same even if you use a different tool. The following content focuses on demonstrating designing primers for different cloning methods.
Exon/Intron Selection
There are many other primer design tools available online, including primer3, and PCR suppliers often offer their own design programs free of charge. Zymo Research’s DNA Clean & Concentrator Kits were designed to facilitate the removal of salts, polymerases, and endonucleases from your PCR product, leaving you with purified, highly concentrated DNA. The resulting DNA is suitable for even the most sensitive downstream applications including sequencing, cloning, ligation, and endonuclease digestion. Discover which PCR purification kit best suits your research goals today. In RT-qPCR, RNA is first converted into cDNA (reverse transcription, or RT), and then amplified with PCR as described in the previous section. During maturation, RNA is subjected to splicing, which removes introns from pre-mRNA sequences.
Manipulating PCR Reagents
That is, as one parameter of PCR is changed, it may impact another. As an example, if the initial PCR was carried out at the sub-optimal annealing temperature (58°C) with an optimal Mg2+ concentration of 2.0 mM, then the result would produce a smear as seen in Figure 3c. An attempt to resolve the smear might involve setting up PCR conditions with reactions containing 2.0 mM MgCl2 and adjusting the annealing temperature to 61°C. Consequently, it is advisable to titrate reagents, rather than adding one concentration to a single reaction, when troubleshooting spurious results.
(a) Self-annealing of primers resulting in formation of secondary hairpin loop structure. Note that primers do not always anneal at the extreme ends and may form smaller loop structures. (b) Primer annealing to each other, rather than the DNA template, creating primer dimers. Once the primers anneal to each other they will elongate to the primer ends.
Primer-BLAST is run on a farm of machines at the NCBI to provide better service for users. Input can be a Genbank accession, a FASTA file, or even primers from another source. It also takes advantage of rich information from NCBI sequence databases to support other requirements such as placing primers based on intron/exon boundaries and SNP locations. Finally, it displays alignments between primers and targets found, allowing the user to make a decision on whether or not to use the primer pairs when potentially unintended targets exist. The Primer-BLAST program consists of a module for generating candidate primer pairs and a module for checking the target specificity of the generated primer pairs. Primer3 is used to generate the candidate primer pairs for a given template sequence.

No comments:
Post a Comment